
No.215, April 2024
Contents
High-performance computing
Licensed software in Slurm (Comsol)
The Slurm workload manager can handle licensed software by assigning available
licenses to jobs at scheduling time. If licenses are not available, jobs are kept
pending until specified licenses become available, rather than failing at runtime
due to a lack of available licenses. This can be particularly useful for software that only has a few
licenses. For example, for the Comsol software, MPCDF currently maintains only two licenses for each
Comsol module for batch use. In order to use this feature, you need to add the -L, --licenses option
to the batch job scripts with a comma-separated list of required license names. To find out
which licenses the mph comsol file requires, you can check it in the comsol GUI or
simply run the mphlicenses command after loading the comsol module on HPC Cobra or
Raven systems:
~> module load comsol/6.2
~> mphlicenses test_file.mph
comsol_nonlinearstructmaterialsbatch@lserv,comsol_comsolbatch@lserv,comsol_structuralmechanicsbatch@lserv,comsol_clusternode@lserv,comsol_comsoluser@lserv
The complete Slurm #SBATCH directive to run this comsol test_file on the clusters then reads
#SBATCH -L comsol_nonlinearstructmaterialsbatch@lserv,comsol_comsolbatch@lserv,comsol_structuralmechanicsbatch@lserv,comsol_clusternode@lserv,comsol_comsoluser@lserv
In order to get a list of all available comsol licenses on HPC systems, run the command:
scontrol show licenses
Mykola Petrov
HPC Software News
Improved workflow for multimer predictions with AlphaFold2 on Raven
AlphaFold2 (AF2) is an artificial-intelligence program released by Google DeepMind which performs predictions of protein structure based on the sequence of amino acids. AF2 has been provided by MPCDF since summer 2021 on Raven and on several institute clusters, and is regularly updated and customized to achieve optimal performance on the MPCDF systems.
Initially, the prediction of the structure of single proteins (monomers) has been a main application of AF2, however more recently the users’ focus shifted towards predicting protein-protein complexes (multimers). As the prediction of multimers is considerably more expensive than most monomer cases, users sometimes hit the 24-hour job time limit on Raven with the original AF2 program, which internally loops over multiple models and random seeds.
To overcome this limitation, MPCDF has refactored the internal double loop over the models and predictions into individual Slurm jobs. In particular, individual Slurm array jobs are now used for each model, each array containing a configurable number of tasks calculating individual predictions using individual random seeds. As a result, each individual prediction task now has a maximum wall clock time of 24 hours.
On Raven, run module help alphafold/2.3.2-2024 to get further instructions.
For multimer cases, copy, adapt and run the script submit_alphafold_jobs.sh
to submit the job-array-based predictions. Users are encouraged to approach us
via the helpdesk with their feedback, which is valuable to further improve this
service.
Klaus Reuter
New version of Intel oneAPI with deprecation of ifort compiler
Intel oneAPI version 2024.0 has been installed on Raven and other HPC
clusters recently. With this installation, we drop the .x suffix and the
patch-level version from the compiler module name. Thus, the Intel compiler
module is now called intel/2024.0. Bugfixes provided by Intel incrementing
the patch level number will be installed under the hood without further
notification. Run module show intel/2024.0 in order to see which exact compiler
version is present. The corresponding MPI module is impi/2021.11, and the latest
MKL module is mkl/2024.0.
As announced previously, MPCDF, starting with intel/2024.0, is using ifx as the default Intel Fortran compiler, together with its MPI wrapper mpiifx. The “classic” Fortran compiler ifort is still present, but is now formally tagged as deprecated by Intel.
Tobias Melson
CUDA modules on Raven
MPCDF offers various CUDA modules on Raven. It depends on the desired compiler,
which of these modules should be picked. If an application code is compiled with the GNU
compiler (gcc/11 or gcc/12), the CUDA modules cuda/* are suitable. In
case the code is compiled with the Nvidia compiler (nvhpcsdk/23), the
matching CUDA modules are named cuda/*-nvhpcsdk (note the suffix here). The
following table lists the compatible CUDA modules for the recent GNU and Nvidia
compilers:
| Compiler | Compatible CUDA modules |
|---|---|
| gcc/11 | cuda/11.4, cuda/11.6 |
| gcc/12 | cuda/12.1, cuda/12.2 |
| nvhpcsdk/23 | cuda/11.8-nvhpcsdk, cuda/12.3-nvhpcsdk |
As announced previously,
CUDA-aware OpenMPI (openmpi_gpu/4.1) is available after having loaded one valid combination of
compiler and CUDA modules.
Tobias Melson, Tilman Dannert
New AMD-GPU ELPA release
The ELPA library provides highly optimized, scalable solvers for generalized and standard, dense symmetric (Hermitian) eigenproblems. Since 2022, the ELPA library supports AMD GPUs and on Europe’s first pre-exascale system LUMI (CSC, Finland) ELPA was successfully employed for solving huge standard eigenvalue problems with a matrix size of up to 3.2 million (leading dimension) on more than 8000 AMD MI250x GPUs. In preparation for the new HPC system Viper at the MPCDF we have released a new version 2024.03.001 of the ELPA library which has been further extensively optimized for AMD GPUs. Among others, support of the GPU-to-GPU collective communications library RCCL (the equivalent to Nvidia’s NCCL library) has been implemented. In addition, the GPU implementation of the routines for the generalized eigenvalue problem has been reworked and speedups of up to a factor of 10 have been achieved. The latest version of ELPA is available to all users as a pre-built software package in the module environment of the MPCDF software stack.
Andreas Marek
Kubernetes in the HPC-Cloud
Containers have been widely adopted as a way to develop, distribute, and deploy applications. Kubernetes provides a framework to run containerized applications in a reliable and scalable fashion. Kubernetes is based on cluster management technology developed at Google (cf. Verma, Abhishek, et al.: “Large-scale cluster management at Google with Borg”, Proceedings of the tenth European conference on computer systems, 2015) and donated to the Linux Foundation in 2015. It has enjoyed wide adoption since.
At MPCDF projects can deploy Kubernetes clusters hosted in the HPC-Cloud based on an OpenStack Heat Orchestration Template as provided by the MPCDF Cloud enabling team. The team maintains two templates: one for a small and simple cluster aimed at projects that wish to test and evaluate Kubernetes, and the second providing a production-grade cluster aimed at projects looking to deploy their services and applications using Kubernetes.
Usage
The canonical way to control a Kubernetes cluster is with kubectl. The
credentials required to connect to the cluster are provided on the control
plane nodes set up by the template. The most important feature is that users
define desired states for pods executing their containers and any peripheral
resources in configuration files.
Many software projects publish the configuration states required to run their
products using Helm, the package manager for Kubernetes. Helm charts
define the desired state of many interacting Kubernetes components. Users can
configure charts by setting desired values of chart variables defining the
specific features of their deployment.
In productive deployments, it is imperative to store and track the configuration files of the services orchestrated in Kubernetes. This can be accomplished, for example, using GitLab. In addition, GitLab offers integrations for Kubernetes that deploy the state defined in the configuration files stored in a repository to Kubernetes as defined in the GitLab CI/CD pipelines. Taking advantage of this feature of GitLab provides a concise way to manage applications for teams of administrators and developers.
Function
Kubernetes is best suited for orchestrating long-running services that are implemented as many inter-dependent microservices. It is also possible to run applications on a schedule, or execute single commands in their containers.
Deploying a web service
Here is a configuration file you can apply with kubectl create -f <filename>.yaml
---
apiVersion: apps/v1
kind: Deployment
metadata:
name: svc-demo-deployment
labels:
app: svc-demo-app
spec:
replicas: 2
selector:
matchLabels:
app: svc-demo-app
template:
metadata:
labels:
app: svc-demo-app
spec:
containers:
- name: svc-demo-container
image: jmalloc/echo-server
ports:
- containerPort: 8080
---
apiVersion: v1
kind: Service
metadata:
name: svc-demo
spec:
selector:
app: svc-demo-app
type: LoadBalancer
ports:
- port: 80
targetPort: 8080
protocol: TCP
The first section defines a deployment with a container image that runs the
application, in this case a server that parrots your HTTP requests. The second
section defines a service, exposing the application using a load balancer. Now
running kubectl get all will give you:
NAME READY STATUS RESTARTS AGE
pod/svc-demo-deployment-587b7c9974-4z5wn 1/1 Running 0 66s
pod/svc-demo-deployment-587b7c9974-lz7pp 1/1 Running 0 66s
NAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE
service/svc-demo LoadBalancer 10.101.113.187 XXX.XXX.XXX.XXX 80:32260/TCP 66s
NAME READY UP-TO-DATE AVAILABLE AGE
deployment.apps/svc-demo-deployment 2/2 2 2 66s
NAME DESIRED CURRENT READY AGE
replicaset.apps/svc-demo-deployment-587b7c9974 2 2 2 66s
You can see two pods running the application and the service making the
application available at http://XXX.XXX.XXX.XXX:80. The application can be
removed with kubectl delete -f <filename>.yaml.
Existing applications
Kubernetes on the HPC-Cloud is already used at MPCDF to host a number of prominent applications, for example MoveApps, NOMAD, or GlycoSHIELD.
Frank Berghaus
GitLab: Graphs & Diagrams
In addition to classic Git functionality, MPCDF’s GitLab instance offers a wide range of capabilities around code organisation and project management. In many of these scopes, for example Wiki pages or issues, the user can enter formatted text in Markdown format. After saving, GitLab will render the Markdown annotations which results in a nicely formatted human readable document. Beside standard Markdown annotations, GitLab integrates out-of-the-box libraries for rendering graph structures and diagrams in Markdown texts. In this article, the creation of graphs and diagrams with the help of the Mermaid library is introduced.
Inside any Markdown-formatted document in GitLab, the following code fragment encapsulates a Mermaid sub document:
```mermaid
graph TD
A --> B
A --> C
C --> D
C --> E
E --> A
```
After saving the Markdown document, GitLab would create the following graph from the code above:
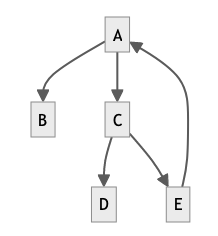
The Mermaid library supports different types of graphs and diagrams. The image below shows some examples, top to bottom: a timeline, a mindmap, a pie chart and - maybe particularly relevant for GitLab users - a Git graph, showing the branch and commit structure of a Git repository:
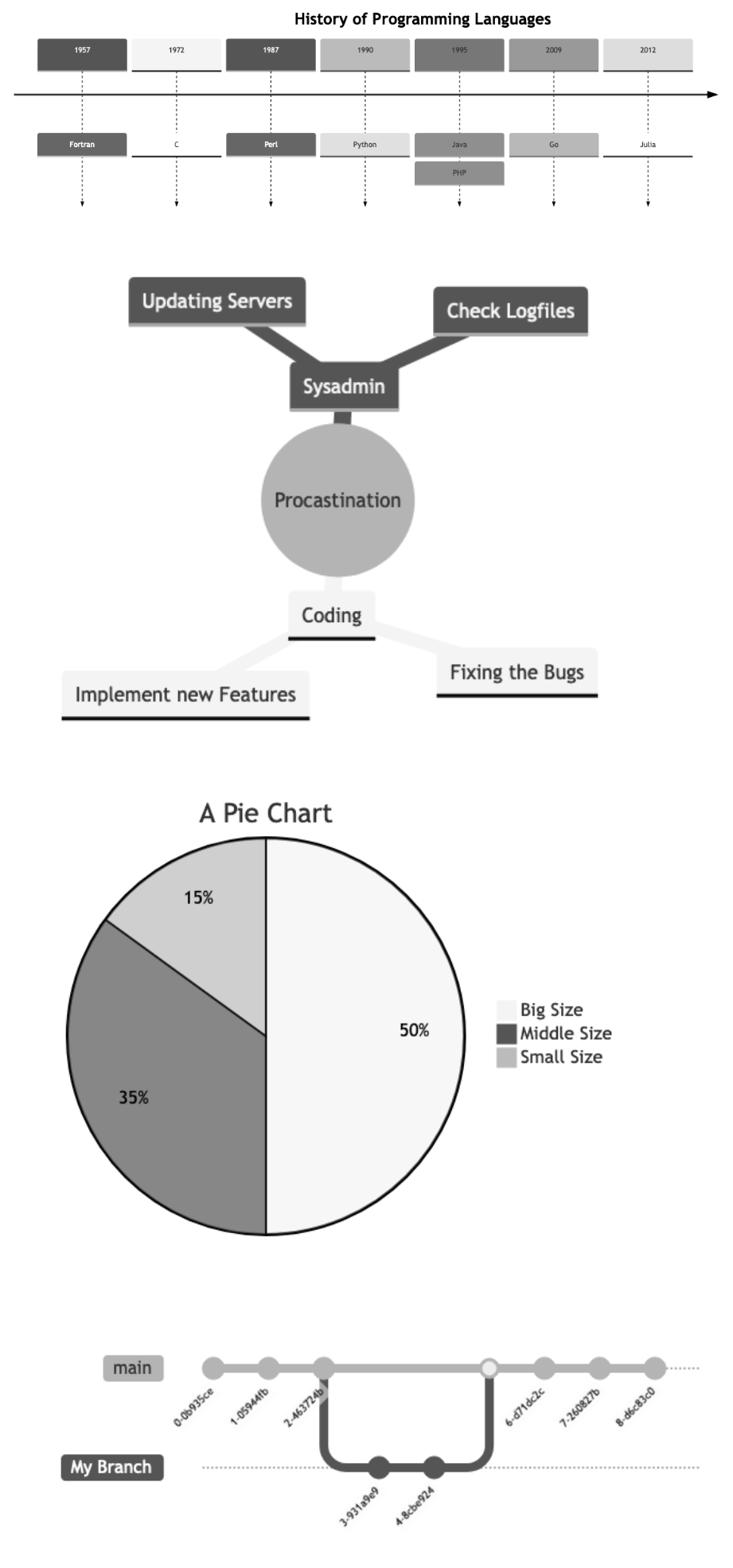
The Markdown and Mermaid code of the examples examples can be found here. Further information about supported graph and diagram types can be found in the Mermaid Documentation.
Alternatives
In the same way GitLab supports the Mermaid library, also PlantUML for UML diagrams and Kroki are supported. And if you don’t want to dive into another descriptive language, GitLab allows you to insert manually drawn diagrams via the external service draw.io. You can find this function as “Insert or edit diagram” button in the Markdown editor (red circle):
Thomas Zastrow
LLMs meet MPCDF

The term “Large Language Model” (LLM), has been part of public discourse since at least the end of 2022, following the release of OpenAI’s ChatGPT. The general-purpose capabilities of these large AI models to process and store unstructured data have ignited a wave of new applications, also in the world of science (cf. The Impact of Large Language Models on Scientific Discovery: a Preliminary Study using GPT-4. Microsoft Research). However, most state-of-the-art models are provided by major tech companies and are accessible only through paid APIs. This paywall, along with the requirement to transfer research data to third parties, often hinders the use of so-called “closed” LLMs. In contrast, “open” LLMs, which can be downloaded and used on-premise, are rapidly narrowing the gap in capabilities compared to their closed counterparts (cf. Open LLM Leaderboard. Hugging Face). However, the immense size and complexity of LLMs require significant computational resources and pose technical challenges for less experienced users, both in terms of operation and training.
Given the computational resources and expertise at the MPCDF, an increasing number of scientists from the Max Planck Society are seeking our advice and support on utilizing LLMs. The inquiries come from a wide range of scientific domains, from natural sciences to the humanities, underscoring the broad impact these models have on the scientific community. The AI group at the MPCDF is actively enhancing its expertise in large language models and expanding its support for various use cases. To this end, we have created the “LLMs Meet MPCDF” GitLab repository, where we start offering examples and guidance on deploying and fine-tuning LLMs on our HPC systems. For now we showcase how to set up an inference server for open models with TGI, and how to supervise-fine-tune a 70B Llama2 model by means of FSDP.
If you have unaddressed use cases or projects involving LLMs, please don’t hesitate to reach out to us, we would be more than happy to hear from and collaborate with you.
David Carreto Fidalgo, Andreas Marek
News & Events
Introduction to MPCDF services
The next issue of our semi-annual online course “Introduction to MPCDF services” will be held on April 25th, 14:00-16:30 via Zoom. Topics comprise login, file systems, HPC systems, the Slurm batch system, and the MPCDF services remote visualization, Jupyter notebooks and datashare, together with a concluding question & answer session. Basic knowledge of Linux is required. No registration is required, just connect at the time of the workshop via the zoom link.
Meet MPCDF
The next editions of our monthly online-seminar series “Meet MPCDF” are scheduled for
May 2nd, 15:30 “CMake - Cross-supercomputer Make” by Vedran Miletic (MPCDF)
June 6th, tba
All announcements and material can be found on our training webpage.
We encourage our users to propose further topics of their interest, e.g. in the fields of high-performance computing, data management, artificial intelligence or high-performance data analytics. Please send an E-mail to training@mpcdf.mpg.de.
Tilman Dannert